Myeloid-NDC

Why settle for less
when you can have it all?
Our first 2024 product, the Myeloid-NDC assay is intended for the integrated and qualitative detection of chromosomal translocations, copy number variants (CNV) and sequence variants from suspected or confirmed cases of myeloid malignancies. The Myeloid-NDC assay has been validated for use with high molecular weight (HMW) genomic DNA (gDNA).
The Myeloid-NDC assay has been designed as an integrated tool covering most genomic alterations recommended for the analysis of acute myeloid leukaemias (AML), myelodysplastic syndromes (MDS) and myeloproliferative neoplasms (MPN). The Myeloid-NDC assay provides a simple solution to allow laboratories to produce integrated reports according to the newest guidelines and recommendations, using just a single DNA sample (with no need for an RNA sample).
Request a quotation
Composition of Myeloid-NDC assay
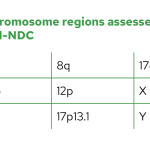
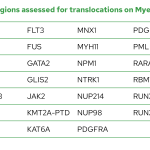
The Myeloid-NDC assay contains most guidelines-recommended genomic regions covering chromosomal translocations and fusion genes, copy number variants (CNV) and sequence variants from suspected or confirmed cases of AML, MDS and MPN.
List of genes with select regions for mutation assessment on Myeloid-NDC
| Gene Name | Region Covered |
|---|---|
| ABL1 | All coding |
| ANKRD26 | Exon 1 |
| ARAF | Exons 4, 7, 10, 15 |
| ASXL1 | All coding |
| ASXL2 | Exons 9-13 |
| ATG2B | Exon 1 |
| ATRX | All coding |
| BCOR | All coding |
| BCORL1 | All coding |
| BRAF | Exons 11, 12, 15 |
| CALR | Exons 8, 9 |
| CBL | Exons 4-9 |
| CBLB | Exons 8-17 |
| CCND2 | All coding |
| CCND3 | Exon 5 |
| CDKN1B | All coding |
| CDKN2A | All coding |
| CDKN2B | All coding |
| CDY1B | Exon 1 (partial) |
| CEBPA | All coding |
| CHEK2 | All coding |
| CREBBP | All coding |
| CSF3R | Exons 14-17 |
| CSNK1A1 | All coding |
| CTCF | Exons 3 (partial), 4-8 |
| CTNNA1 | Exons 6-17 |
| CUX1 | All coding |
| DDX41 | All coding |
| DHX15 | Exons 2-5 |
| DIS3 | All coding |
| DNMT3A | Exons 4-23 |
| EP300 | All coding |
| ERBB3 | Exons 3-8, 15, 22-24 |
| ETNK1 | Exon 3 |
| ETV6 | All coding |
| EZH2 | All coding |
| FBXW7 | All coding |
| FLT3 | Exons 11, 13-15, 16, 20 |
| GATA1 | All coding |
| GATA2 | All coding |
| GATA3 | Exon 6 |
| GNAS | All coding |
| GNB1 | Exons 5-6 |
| HRAS | Exons 2-4 |
| IDH1 | Exon 4 |
| IDH2 | Exon 4 |
| IKZF1 | All coding |
| JAK2 | Exons 8-19 |
| JAK3 | Exons 11-22 |
| KDM6A | All coding |
| KDR | Exons 11, 12, 23 |
| KIT | Exons 8-11, 13, 17-18 |
| KMT2A | Exons 6-27 |
| KMT2C | All coding |
| KMT2E | All coding |
| KRAS | Exons 2-4 |
| MAP2K1 | Exons 2-3 |
| MAP3K1 | Exons 14-20 |
| MAPK1 | Exons 4, 7 |
| MBD4 | All coding |
| MGA | Exons 3, 9, 18 |
| MPL | Exons 10-12 |
| MST1 | Exons 7-14 |
| MYC | Exon 2 |
| NF1 | All coding |
| NFE2 | All coding |
| SRP72 | Exons 6-10 |
| SRSF2 | Exon 1 |
| STAG2 | All coding |
| STAT5B | Exons 14-16 |
| TERC | Exon 1 |
| TERT | All coding and promoter |
| TET2 | All coding |
| NPM1 | Exons 10-11 |
| NRAS | Exons 2-4 |
| NSD1 | All coding |
| NUP98 | All coding |
| PHF6 | All coding |
| PIK3CA | Exons 10, 21 |
| PIK3CD | Exons 9-11 |
| PML | Exon 6 |
| PPM1D | Exon 6 |
| PTPN11 | Exons 3, 8, 12-13 |
| RAD21 | All coding |
| RB1 | All coding |
| RIT1 | Exon 5 |
| RUNX1 | Exons 4-9 |
| RUNX1T1 | All coding |
| SAMD9 | Exon 3 |
| SAMD9L | Exon 5 |
| SETBP1 | Exon 4 |
| SF3B1 | Exons 13-18 |
| SH2B3 | Exons 2 (partial), 3-8 |
| SMC1A | All coding |
| SMC3 | All coding |
| TP53 | Exons 2-10 |
| U2AF1 | Exon 2 |
| UBA1 | Exon 3 |
| WT1 | All coding |
| ZBTB7A | Exon 2 |
| ZRSR2 | All coding |
Performance specifications of the Myeloid-NDC
Expected Metrics:
The table below shows the approximate expected metrics of the Myeloid-NDC assay based on 16 samples per hybridisation reaction (plus controls) and run on a NextSeq500/550 (Mid- Output 150 cycles cartridge) at 2 x 75bp. These can vary depending on DNA quality and quantity, technical factors in each laboratory, differences in equipment, performance of libraries, etc.
| Parameter | Expected value |
|---|---|
| Library yield (ng) | 3,000 (range: 1,000-6,000) |
| Library average Library average fragment size (bp) | 330 (range: 275-450) |
| Average on/near target bases (%) | 80% (range: 70%-85%) |
| Duplicate reads (%) | 25% (range: 15%-35%) |
| Mean target unique coverage (reads) | 1,000x (range: 500x-1,500x) |
Observed Analytical Performance:
The following table represents the observed analytical performance for the Myeloid-NDC assay:
| Sensitivity | Specificity | Limit of Detection | |
|---|---|---|---|
| SV | >95% | >99% | 5% |
| VAR | >99% | >99% | 4% |
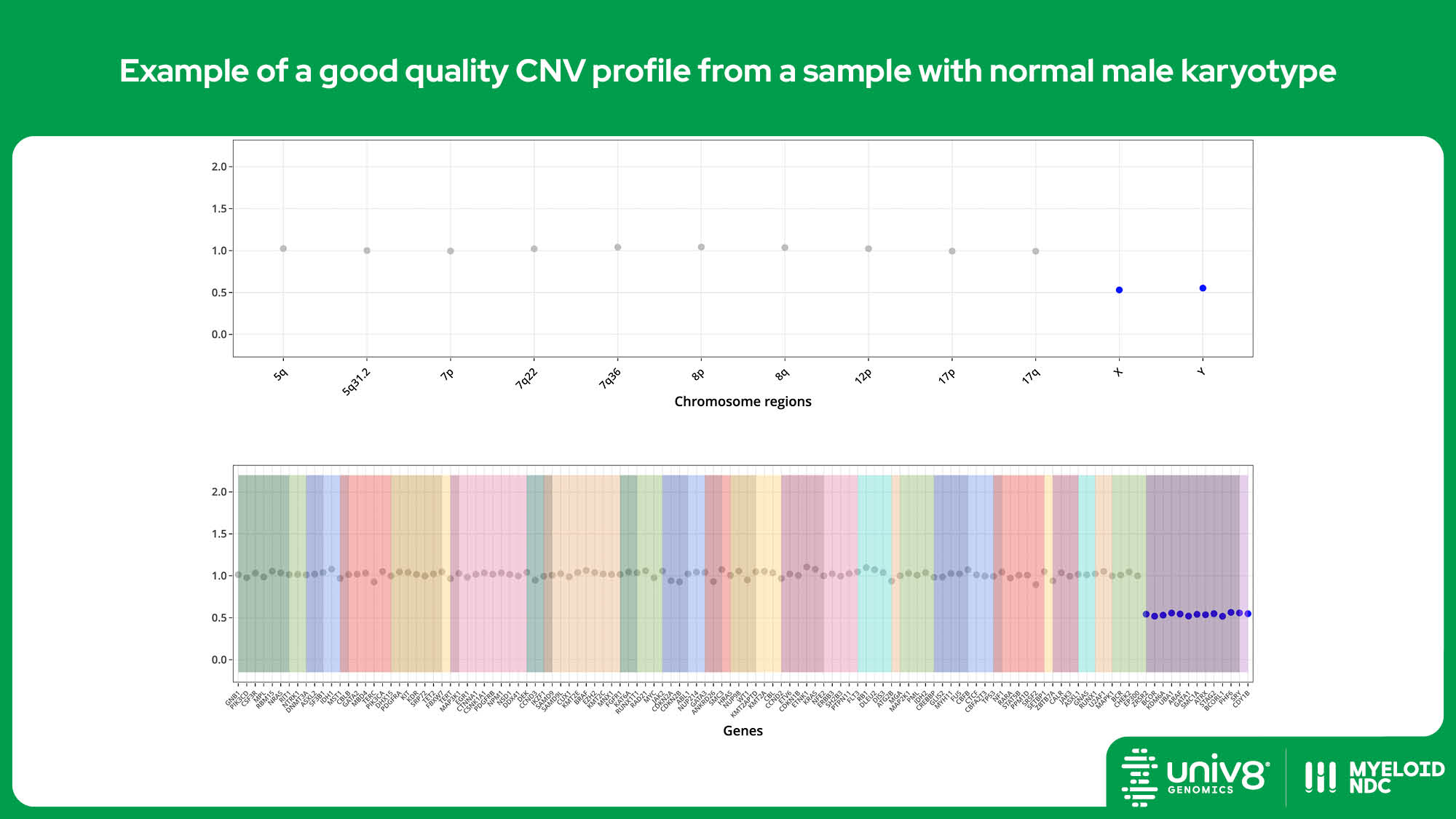
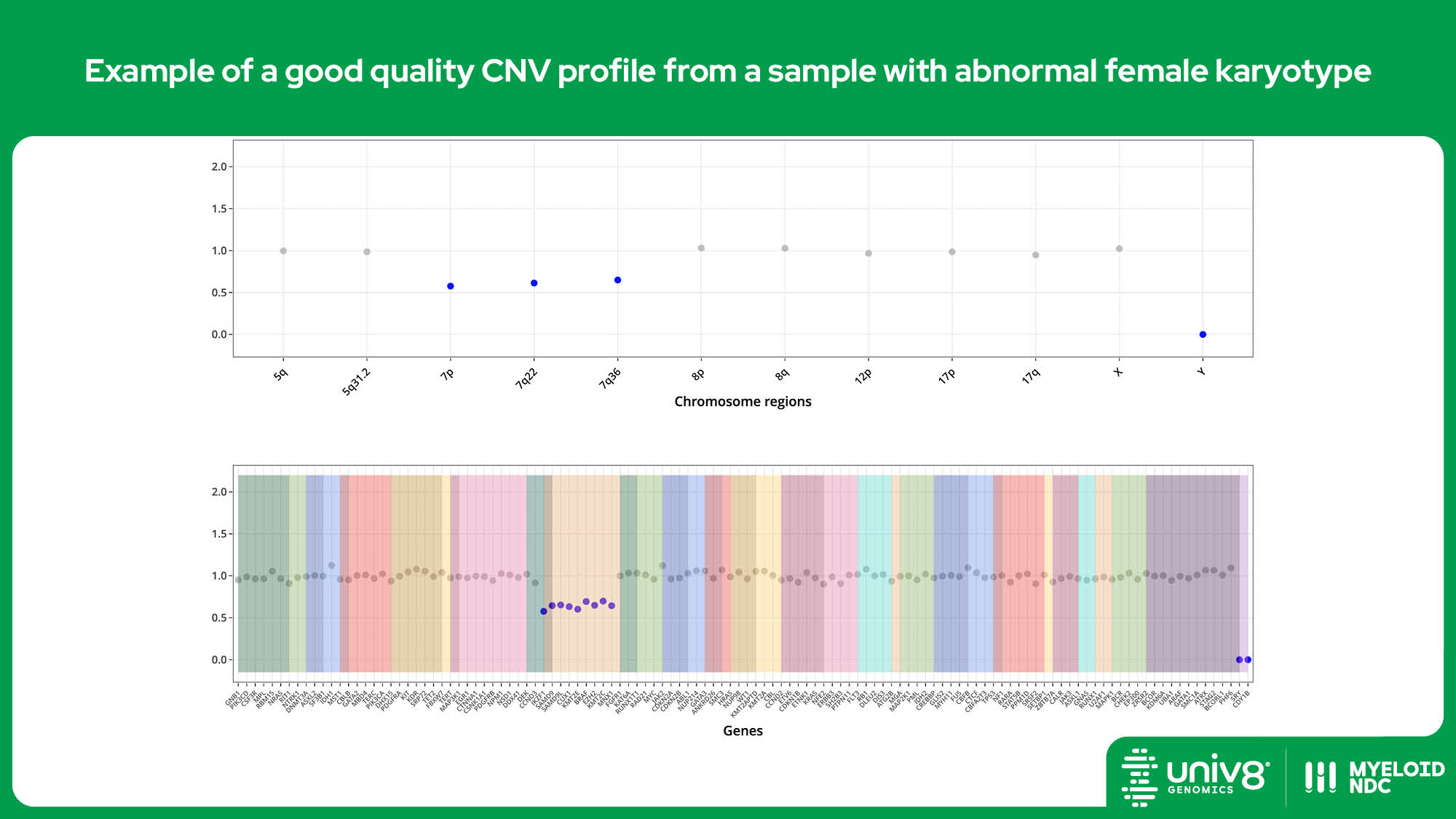
| CNV* | >95% | >97% | 40% (tumour cells) |
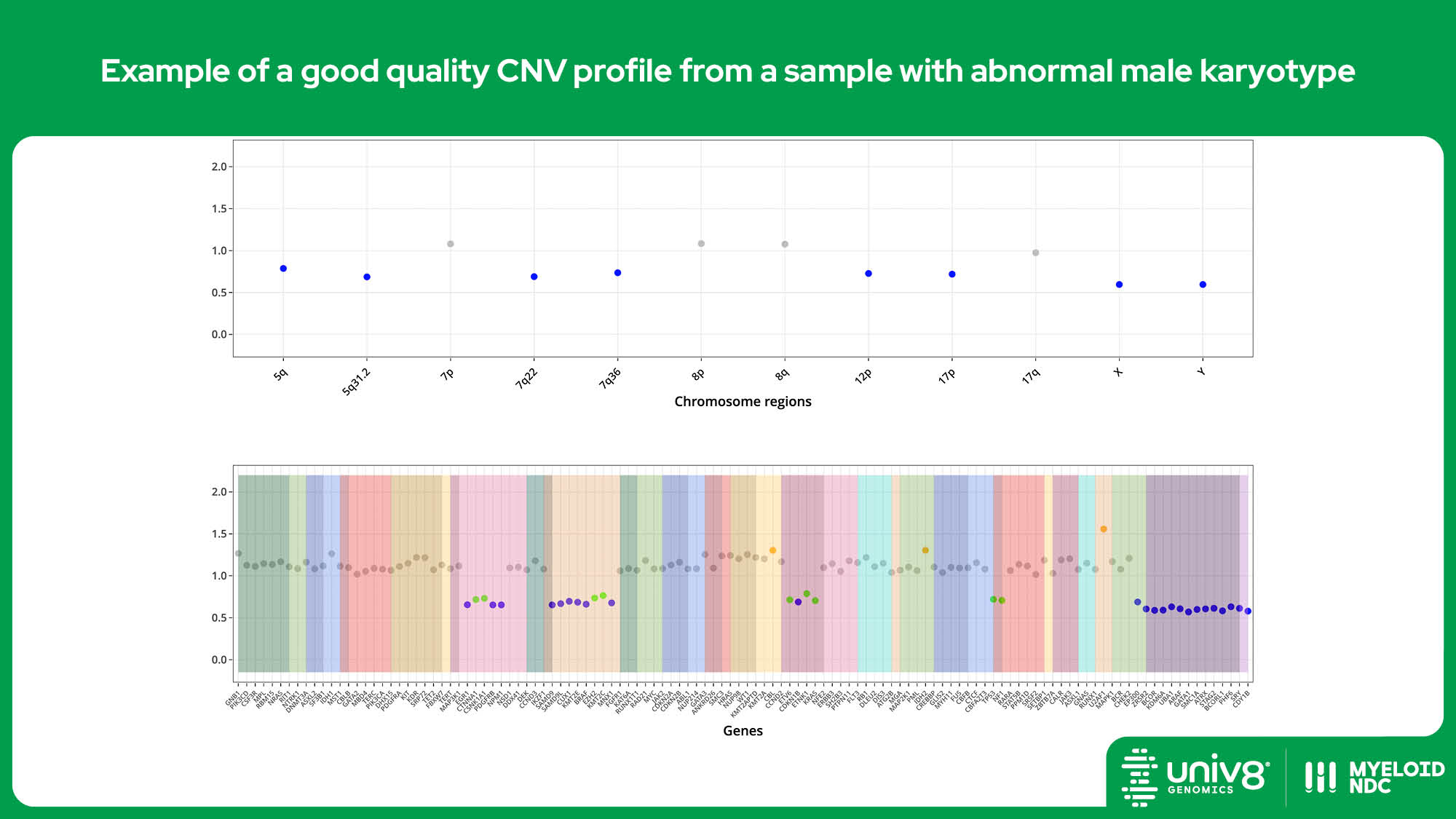
SV: structural variants, VAR: sequence variants, CNV: copy number variants. *The threshold baseline fold-change for deletions detected by Myeloid-NDC is 0.8 and 1.3 for losses and gains, and 0.7 and 2.0 for deletions and amplifications, respectively.