EuroClonality-NDC


Why settle for less
when you can have it all?
Our first product, the EuroClonality-NDC assay allows you to detect immunoglobulin (IG) and T-cell receptor (TR) rearrangements and translocations, as well as mutations and copy number alterations in clinically relevant genes for most lymphoproliferative disorders.
EuroClonality-NDC has been designed and developed by the EuroClonality-NGS working group (an independent subdivision of EuroClonality) and validated in a multi-centre international study involving expert clinical and academic laboratories across Europe.

Request a quotation
What our customers say
Composition of EuroClonality-NDC assay
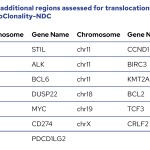
The EuroClonality-NDC assay contains a series of genomic regions covering all the IG (including IGH switch regions) and TR genes, intronic regions of some commonly translocated genes, coding regions of clinically-relevant genes and specific regions for copy number variation (CNV) analysis.
List of genes with full coding regions mutation assessment on EuroClonality-NDC
| ARID1A | EP300 | MAP3K14 | SAMHD1 |
| ATM | ERG | MAPK1 | SOCS1 |
| B2M | FAT1 | MYC | TCF3 |
| BCL2 | H1-5 | PAX5 | TENT5C |
| BTG1 | H1-2 | PHF6 | TET2 |
| BTK | H1-3 | POT1 | TNFAIP3 |
| CDKN2A | H1-4 | PTEN | TP53 |
| CDKN2B | ID3 | PTPRD | TRAF2 |
| CDKN2C | KLF2 | RHOA | TRAF3 |
| CKS1B | KMT2D | RUNX1 | WT1 |
List of genes with select regions for mutation assessment on EuroClonality-NDC
| Gene Name | Region Covered |
|---|---|
| ABL1 | Exons 4-9 |
| ASXL1 | Hotspot in last exon |
| BIRC3 | Exons 8 and 9 |
| BRAF | Exons 11 and 15 |
| CARD11 | Exons 2-16 |
| CCND1 | Exon 1 |
| CCND3 | Exon 5 |
| CD79A | Exons 4 & 5 |
| CD79B | Exons 4-6 |
| CREBBP | Exons 2-30 |
| CXCR4 | Hotspot in exon 2 |
| DIS3 | Exons 1-3 and 9-18 |
| DNMT3A | Exons 8-23 |
| EGR2 | Hotspot in exon 2 |
| EZH2 | Exons 15-17 |
| FBXW7 | Exons 1-12 |
| FOXO1 | Exon 1 |
| IDH1 | Exon 4 |
| IDH2 | Exons 4 and 7 |
| IKZF1 | Exons 2-7 |
| IL7R | Exon 6 |
| IRF4 | Exons 2-3 |
| JAK1 | Exons 12-15 |
| JAK2 | Exons 12-15 |
| JAK3 | Exons 11-24 |
| KIT | Exons 9, 11, 13, 17 and 18 |
| KRAS | Exons 2, 3 & 4 |
| MAP2K1 | Exons 1-8 |
| MEF2B | Exons 2-4 |
| MYD88 | Exon 3-5 |
| NFKBIE | Exon 1 |
| NOTCH1 | Exons 26, 27 & 34 |
| NOTCH2 | Exon 34 |
| NRAS | Exons 2-4 |
| NT5C2 | Exons 9-15 |
| PIK3CA | Exons 2, 5 10 & 21 |
| PLCG1 | Exons 1, 11, 15-17, 19 and 29 |
| PLCG2 | Exons 19, 20 & 24 |
| SF3B1 | Exons 12-17 |
| STAT3 | Exons 10-16 and 20-21 |
| STAT5B | Exons 13-19 |
| STAT6 | Exons 5, 9, 11, 12 and 14 |
| XPO1 | Exons 15-16 |
Performance of the EuroClonality-NDC assay
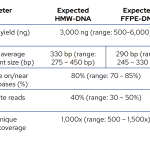
The expected metrics for library preparation and hybridisation performance ensure sufficient coverage of all targets to obtain robust and reliable results.
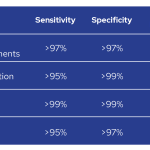
Analytical sensitivity varies according to the type of alterations, from >95% for CNV up to >99% for sequence variants at >4% VAF. Analytical specificity also varies from >97% for CNV and IG/TR rearrangements up to >99% for sequence variants at >4% VAF.
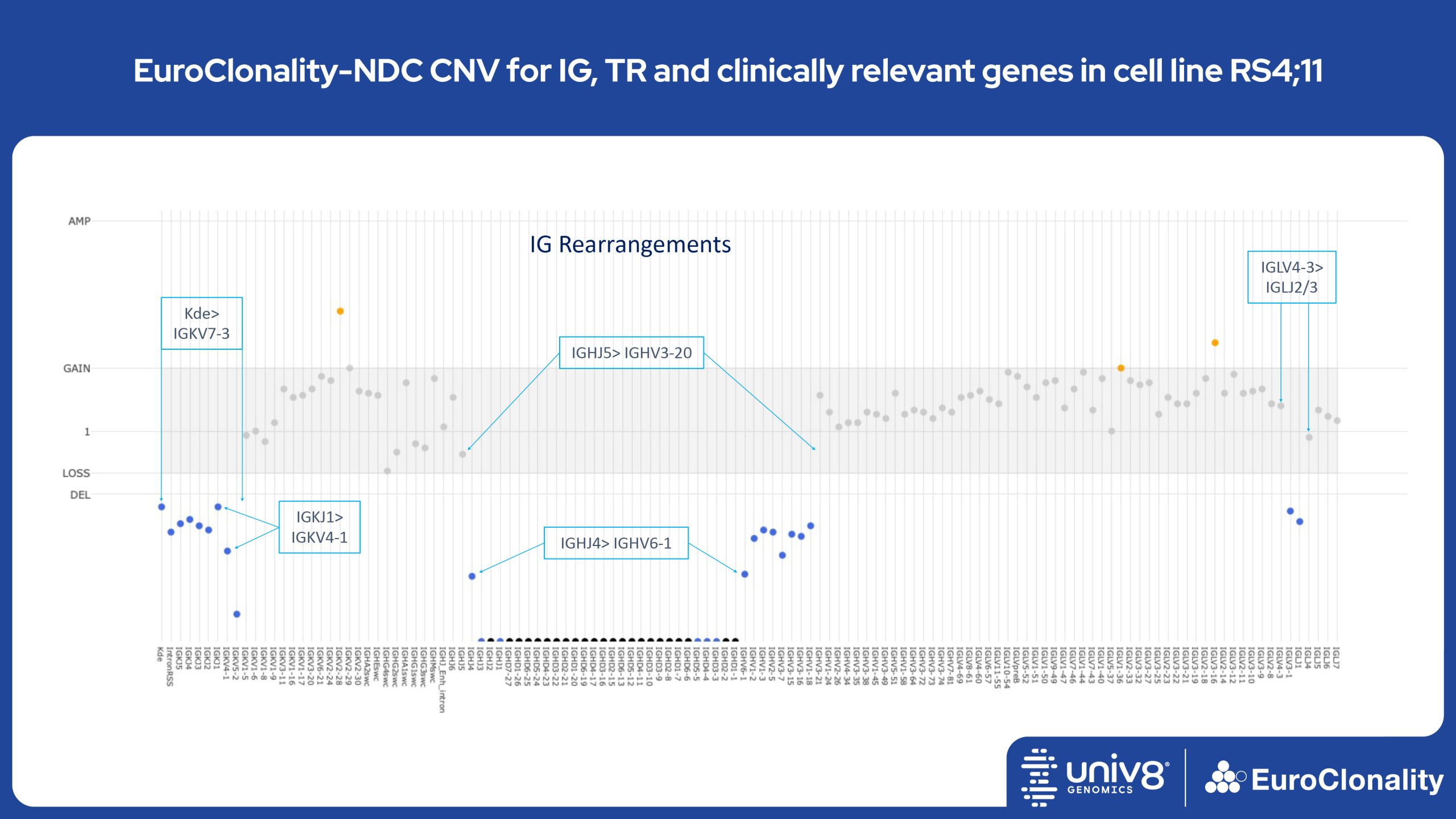
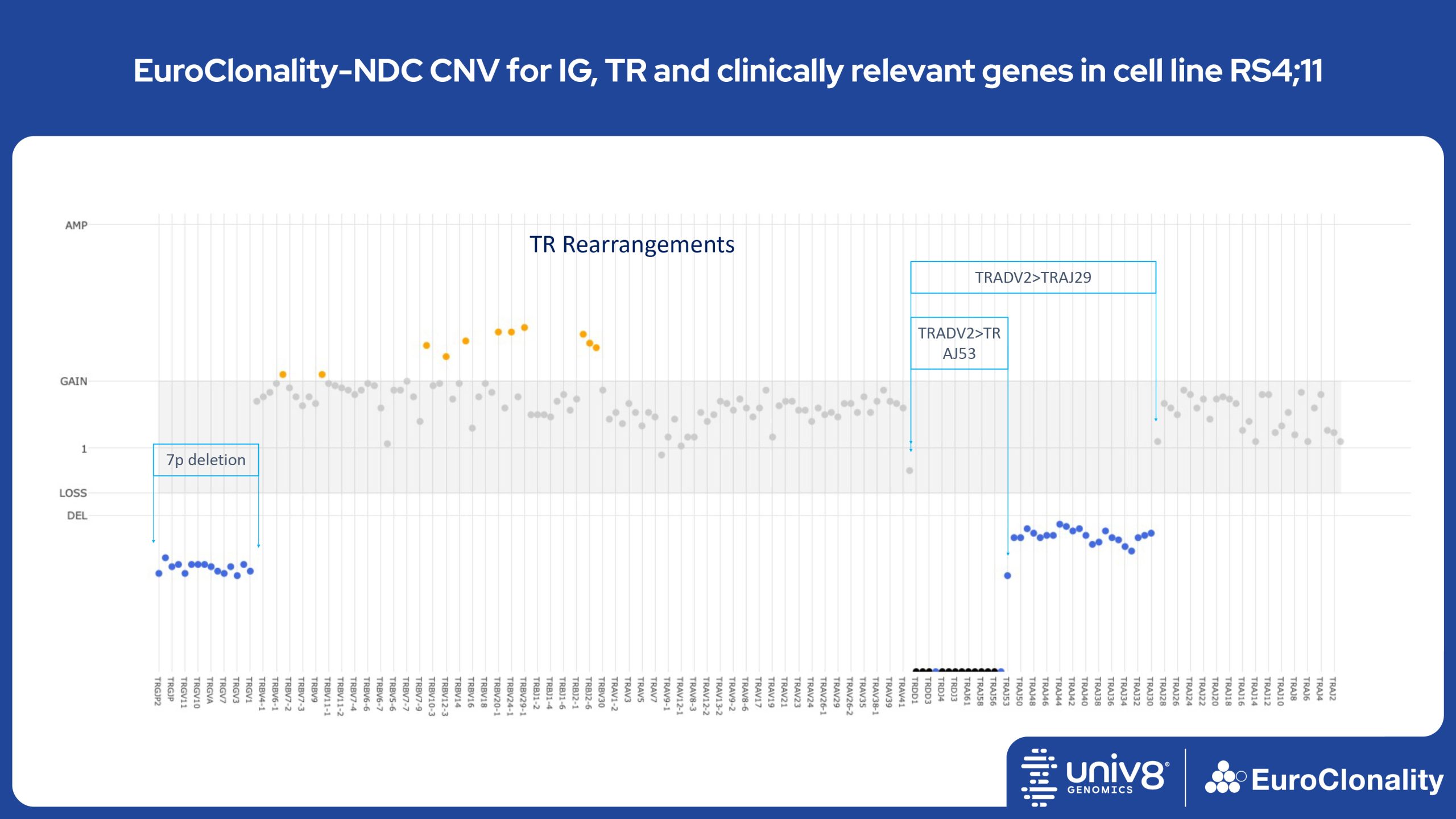
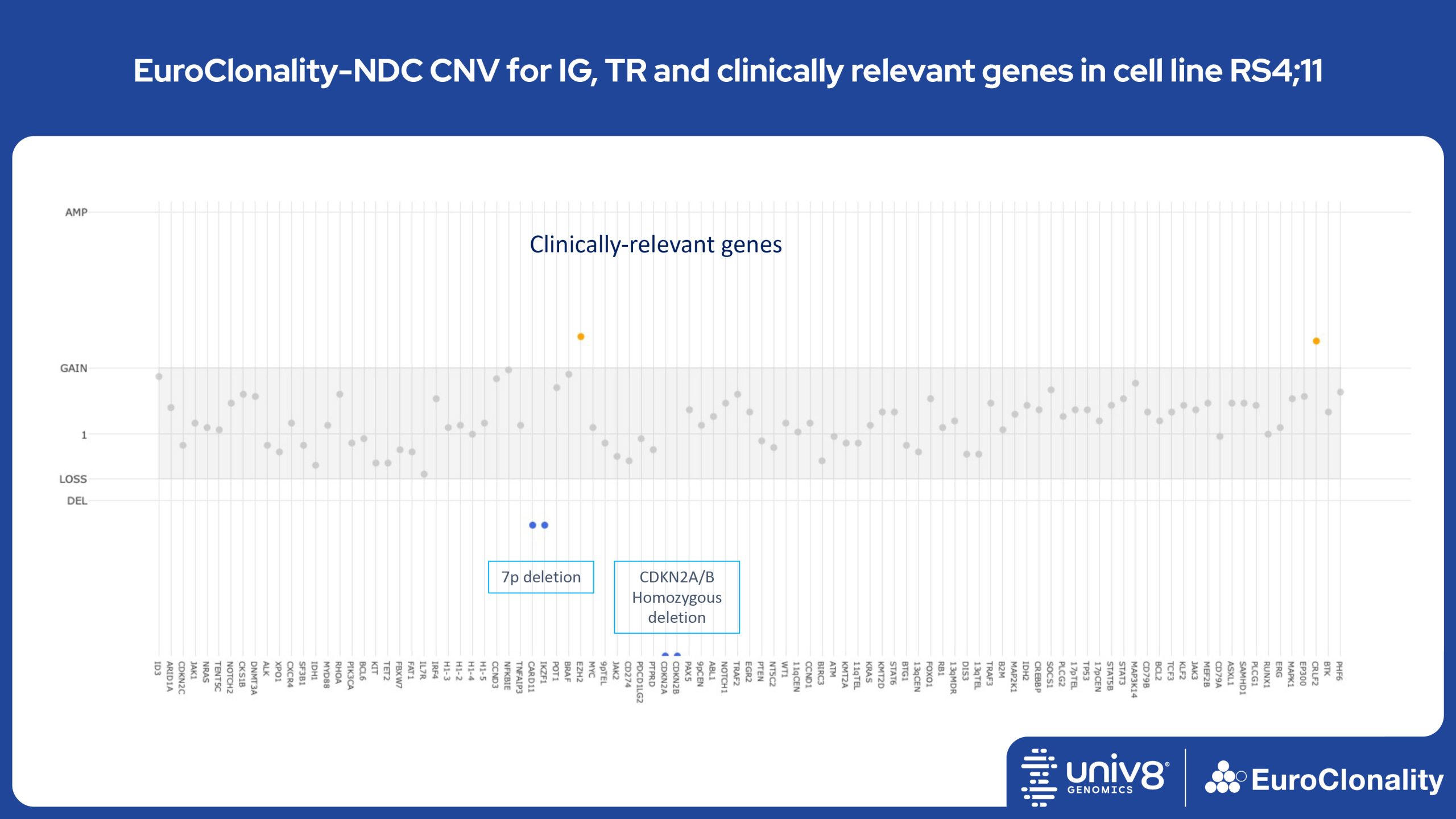
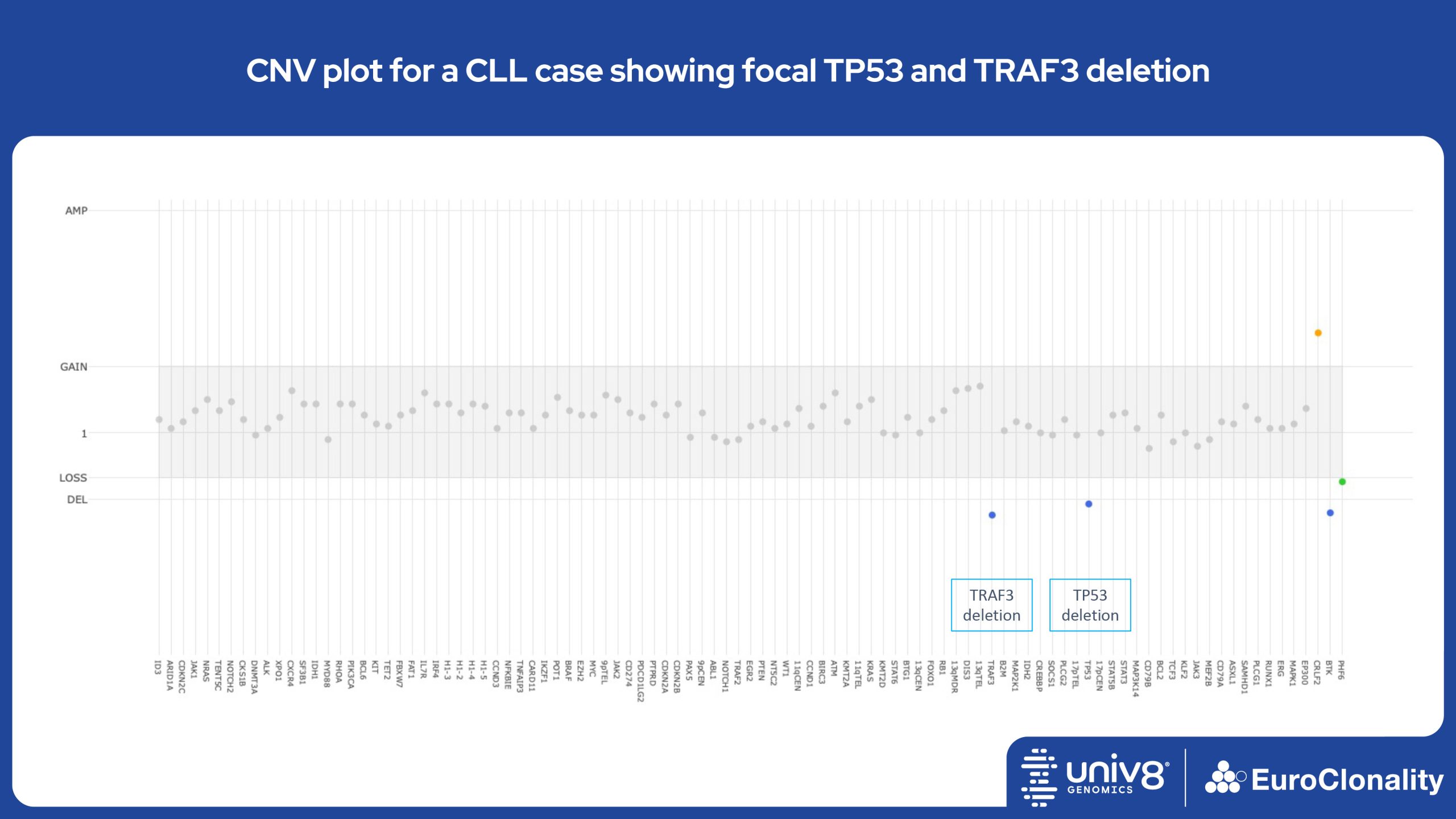
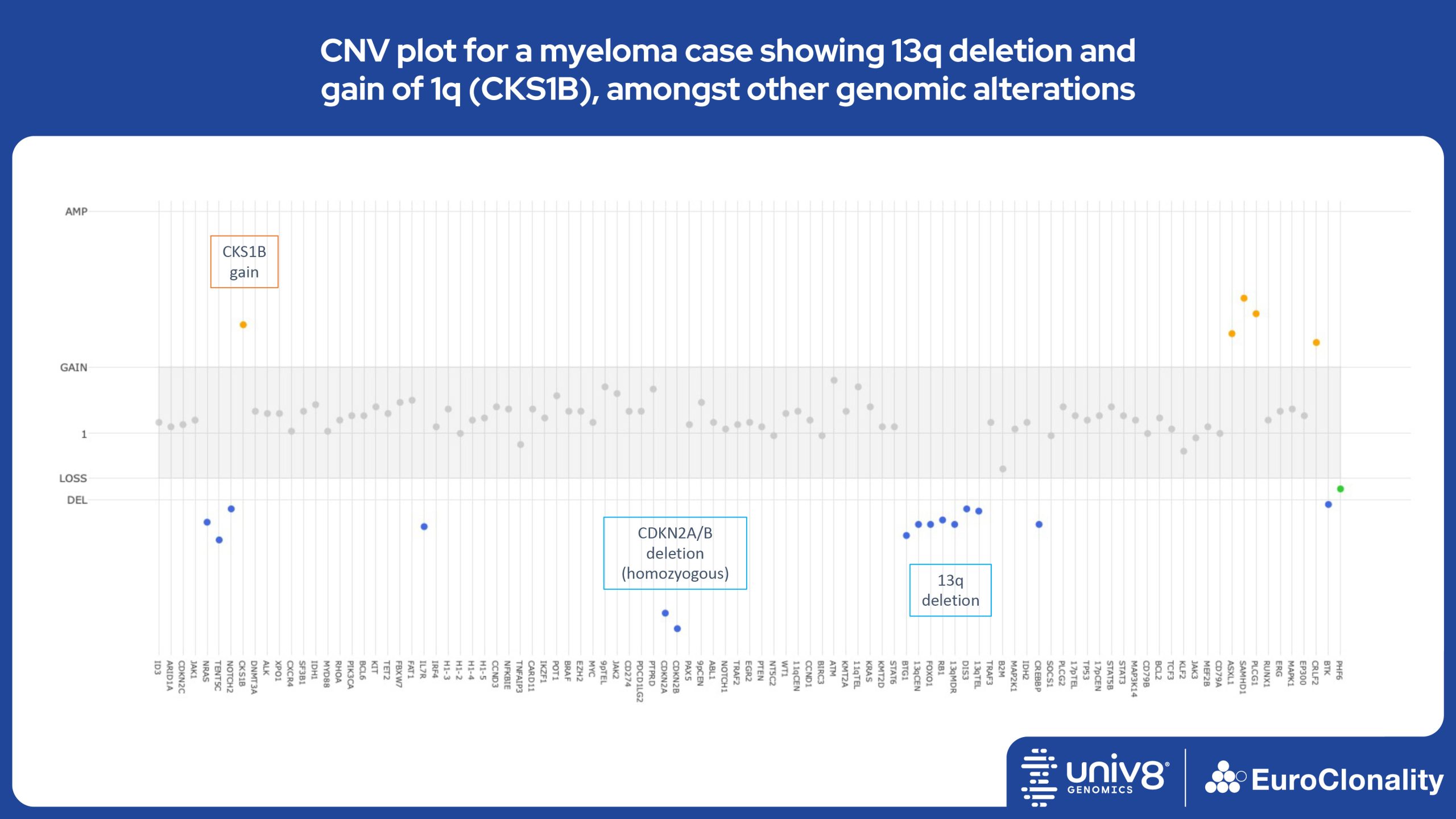
Download Quick Reference and Analysis GuidesEuroClonality-NDC CNV output for IG, TR and clinically relevant genes in cell line RS4;11
The EuroClonality-NDC assay produces copy number variant (CNV) outputs specifically for IG and TR rearrangements that can be used alongside the Rearrangement results to assess clonality in an orthogonal fashion. These CNV plots are most helpful when the tumour content is at least 40%. There is also an additional output (called ’Onco’) that shows CNV data across a range of clinically relevant genes and genomic regions included in the assay. CNV data have been validated for high molecular weight DNA samples containing more than 40% tumour cells.
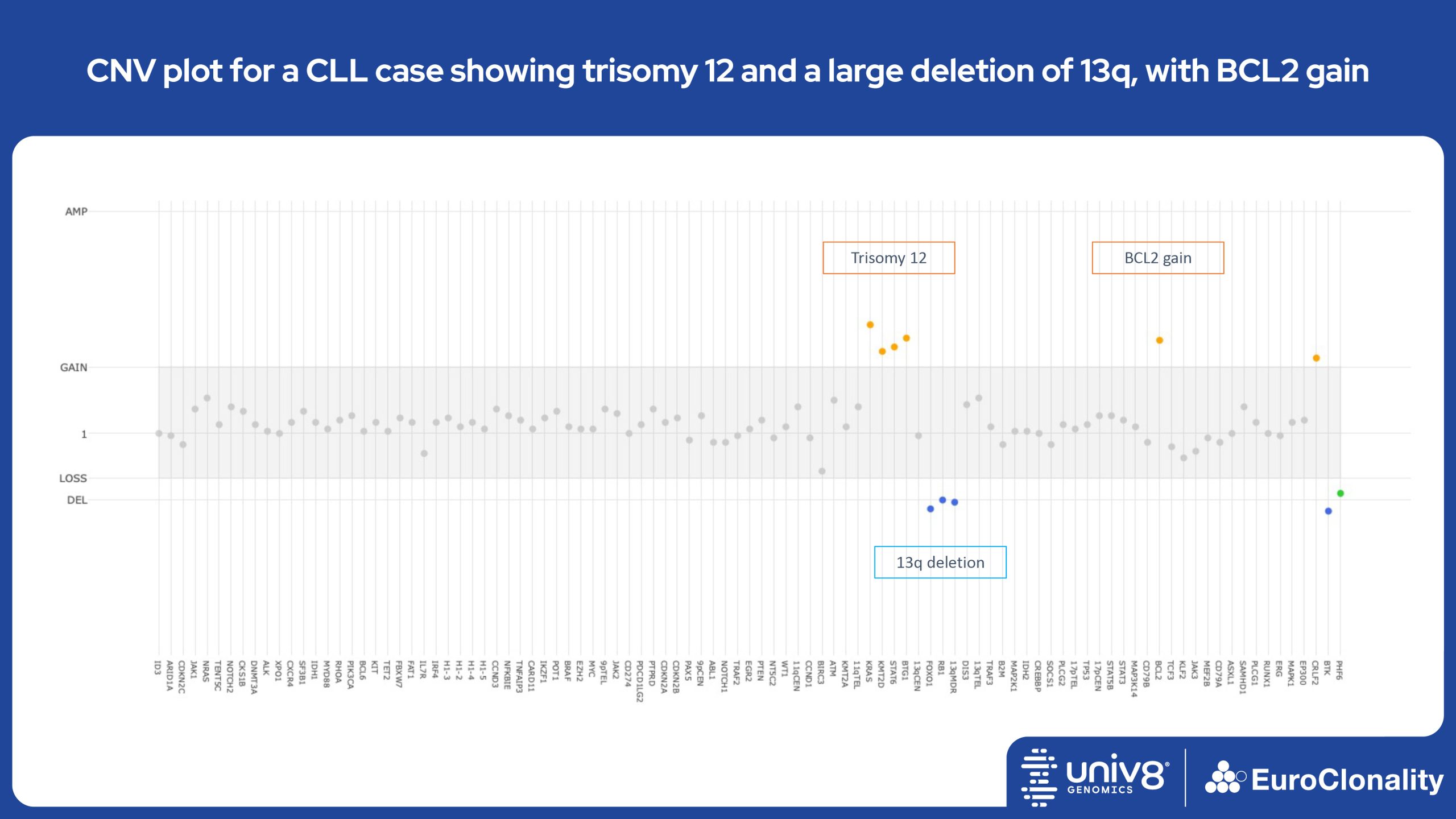
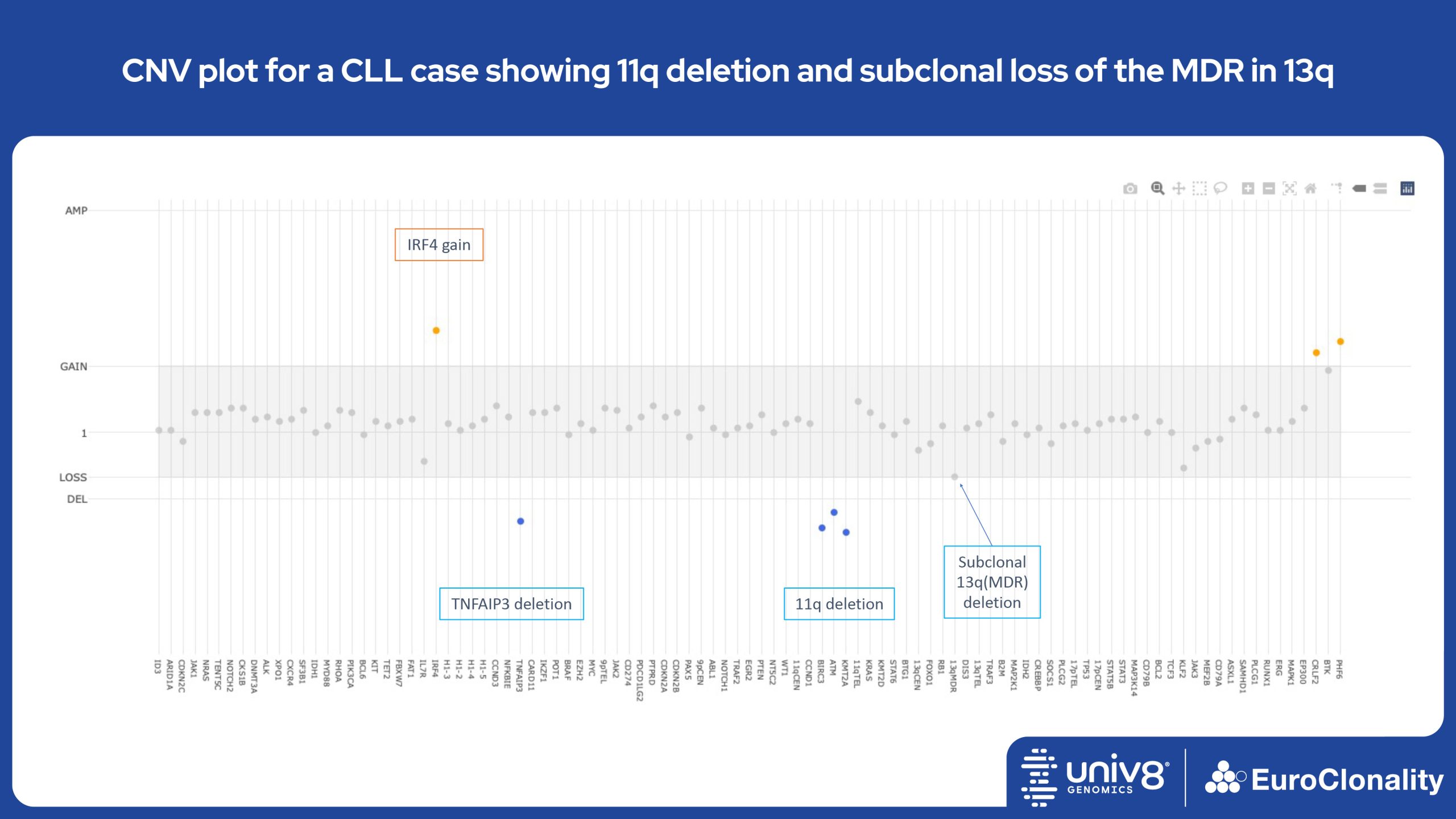
Download examples of results produced by the EuroClonality-NDC assayEuroClonality-NDC Copy number alteration plots in clinical samples
Clinical and academic centres involved in the EuroClonality-NDC validation
| Queen’s University Belfast | Belfast, UK |
| Universitätsklinikum Schleswig-Holstein (UKSH) | Kiel, GERMANY |
| CEITEC – Central European Technology Institute | Brno, CZECH REPUBLIC |
| The Royal Marsden NHS Trust | London, UK |
| Centro Maria Letizia Verga | Monza, ITALY |
| Hospital Universitario de Salamanca | Salamanca, SPAIN |
| Hopital Necker-Enfants Malades | Paris, FRANCE |
| Erasmus MC, university medical center Rotterdam | Rotterdam, THE NETHERLANDS |
| Centre for Research and Technology Hellas | Thessaloniki, GREECE |
| University of Torino | Torino, ITALY |
| Radboud university medical centre Nijmegen | Nijmegen, THE NETHERLANDS |
| Hopital Pitié-Salpétrière | Paris, FRANCE |